While GeNIe structure learning algorithms do not allow for learning the structure of dynamic models, it is possible to learn the parameters of existing DBNs from time series.
To learn parameters of an existing DBN (i.e., one for which the structure is already defined), you will need both, a data file and a network open. We will demonstrate the procedure of learning the parameters of a DBN from data on the network created in the section Creating DBNs and a corresponding data file, both available in the example models directory. Here is a screen shot of the data file opened in GeNIe:
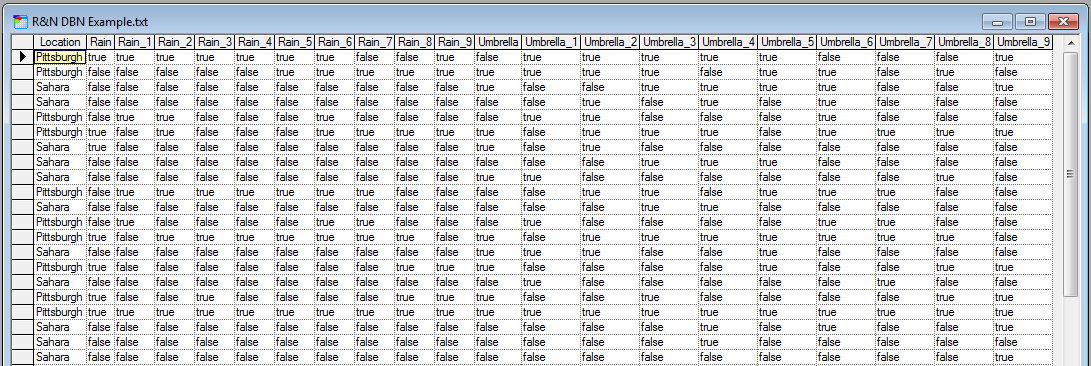
The file used for learning the parameters of a DBN has to follow a simple but strict format. The labels have to correspond to the IDs of the nodes in the network. Measurements taken at different time steps have to be labeled by the node ID with an underscore character followed by the time step number (except for time step zero, which has no time step mark).
Once you have opened both the model and the data, select Learn Parameters... from the Data menu.
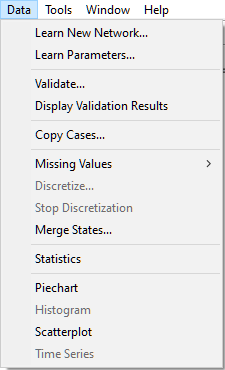
This will invoke the Time series columns dialog that allows for double-checking the matching between time steps and labels in the data set.
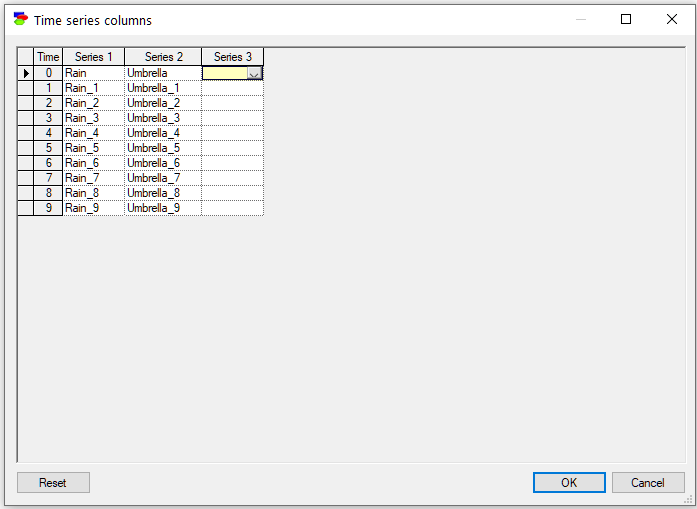
Closing this dialog invoke the Match Network and Data dialog that serves to create a mapping between the variables defined in the network (left column) and the variables defined in the data set (right column).
Both lists of variables are sorted alphabetically. The Match Network and Data dialog does text pre-matching and places in the central column all those variables and their states that match (have identical or close to identical names). If there is any disparity between them, GeNIe highlights the differences by means of a yellow background, which makes it easy to identify disparities. Manual matching between variables in the model and the data is performed by dragging and dropping (both variables and their outcomes). To indicate that a variable (or its state or its state in the middle column) in the model is the same as a variable in the data, simply drag-and-drop the variable (or its state in the middle column) from one to the other column. Please note that it is only necessary to match the variables in the model to the columns that correspond to the time step zero in the data. Other times steps (double-checked in the Time series columns dialog) will follow.
To start the matching process from scratch, use the Reset button, which will result in the following matching:
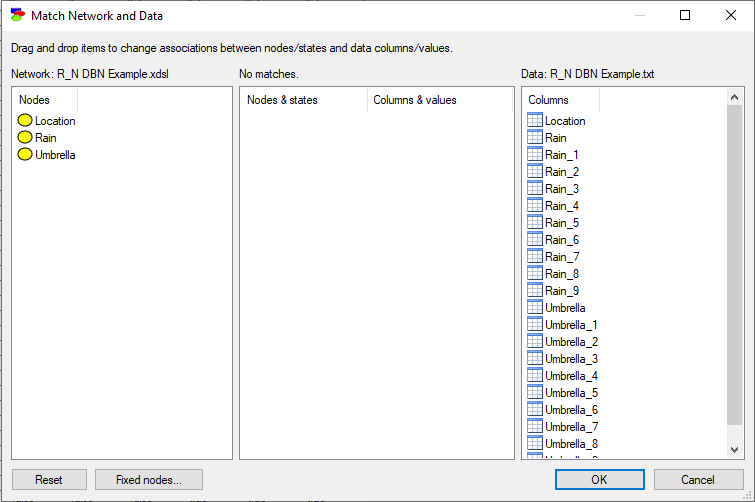
Fixed nodes... button, described in detail in section Learning parameters, invokes a dialog that allows for excluding nodes from the learning process.
Once you have verified that the model and the data are matched correctly, press OK, which will bring up the following dialog:
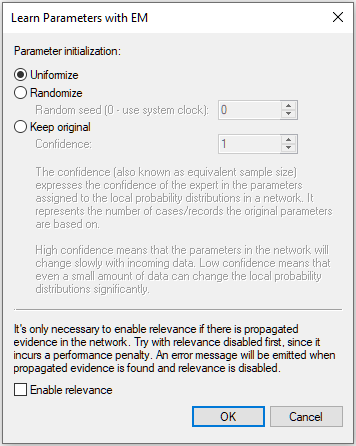
This dialog is described in detail in section Learning parameters.
Once we press OK, the EM algorithm updates the network parameters following the options chosen and comes back with the following dialog:
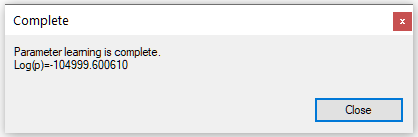
Log(p), ranging from minus infinity to zero, is a measure of fit of the model to the data.
A remark on the network structure and also on existing evidence. Learning parameters functionality focuses on learning parameters, not the structure, which is assumed fixed and will be unaffected. Existing evidence in the network is ignored and has no effect on the learned parameters.