To learn parameters of an existing network (i.e., one for which the structure is already defined), you will need both, a data file and the network open. We will demonstrate the procedure of learning the parameters of a Bayesian network from data on the network Credit.xdsl and a data file Credit10K.csv. Both are available among the example files included with GeNIe. Once you have opened both, select Data-Learn Parameters...
This will invoke the Match Network and Data dialog that serves to create a mapping between the variables defined in the network (left column) and the variables defined in the data set (right column).
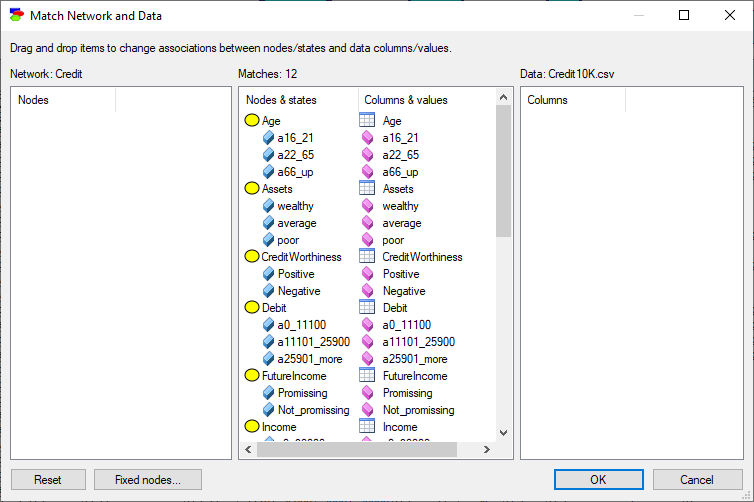
Both lists of variables are sorted alphabetically. The Match Network and Data dialog does text pre-matching and places in the central column all those variables and their states that match (have identical or close to identical names - GeNIe is rather tolerant in this respect and, for example, ignores spaces and special characters). If there is any disparity between them, GeNIe highlights the differing labels by means of a yellow background. Manual matching between variables in the model and the data can be performed by dragging and dropping (both variables and their outcomes). To indicate that a variable (or its state in the middle column) in the model is the same as a variable in the data, simply drag-and-drop the variable (or its state in the middle column) from one to the other column. To start the matching process from scratch, use the Reset button, which will result in the following matching:
Fixed nodes... button invokes a dialog that allows for excluding nodes from the learning process:
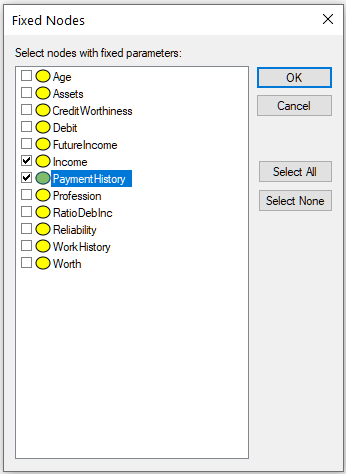
Nodes selected in this dialog (in the dialog above, nodes Income and PaymentHistory) will not be modified by the learning process and will preserve their original CPT.
Once you have verified that the model and the data are matched correctly, press OK, which will bring up the following dialog:
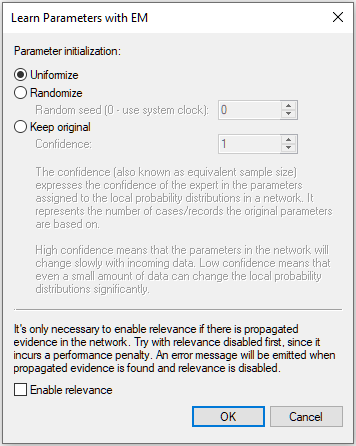
In learning probability distributions, GeNIe uses the EM algorithm (Dempster et al., 1977; Lauritzen, 1995), which is capable of learning parameters from data sets that contain missing values (this is often the case). The algorithm has several parameters:
The Parameter initialization group allows for choosing one of the three possible starting points of the EM algorithm.
Uniformize, when set, causes the algorithm to start with all parameters in the network taken from the Uniform distribution. This is a typical option that one should use when one wants to disregard the existing parameters. The Confidence assigned by the algorithm in this case is equal to 1.
Randomize allows for picking random values for parameters, which inserts some randomness in the algorithm's search for the optimal values of parameters. Interestingly, uniformizing all distributions prior to learning does not necessarily lead to better quality parameters. Any way you set the initial values of the parameters will just provide a starting point in EM's search for the parameter set that maximizes the probability of data given the model. Using the Randomize option may be especially useful when learning parameters with latent variables. The EM algorithm is more likely in such cases to avoid the local maximum around uniform distributions. Random number seed is the initial seed passed to the random number generator. Using the same seed each time you perform learning makes the results perfectly reproducible, unless the seed is equal to zero (the default value), in which case GeNIe uses the system clock as the seed and the random number sequence is really random.
Keep original allows for starting with the original parameters. This option should be used only if we use the new data set as an additional source of information over the existing network. Keeping the original parameters and learning from the same data file that they were extracted from will lead to over-fitting the data.
When keeping the original probabilities in the network (Keep original option), Confidence becomes important. Confidence is also known as the equivalent sample size (ESS) and can be interpreted as the number of records that the current network parameters are based on. The interpretation of this parameter is obvious when the entire network or its parameters have been learned from data - it should be equal to the number of records in the data file from which they were learned. The Confidence in the screen shot below is set to 100.
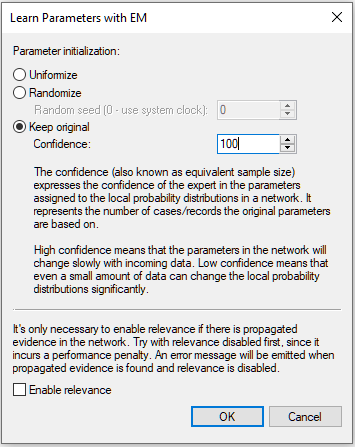
When the parameters in the network have been elicited from an expert, we can view them as the number of cases that the expert has seen before providing us with the current parameters. The larger the ESS, the less weight is assigned to the new cases, which gives a mechanism for gentle refinement of model numerical parameters. ESS expresses the confidence of the expert in the parameters assigned to the local probability distributions in the network. High confidence means that the parameters in the network will change slowly with incoming data. Low confidence means that even a small amount of data can change the local probability distributions significantly. In establishing a value for ESS, we advise to reflect on the number of records/cases on which the current parameters are based. This will naturally combine with the number of new records, a quantity that is known by the algorithm in the learning process.
Enable relevance option makes the algorithm faster by speeding up the Bayesian inference part. We suggest that this be checked only in the rare cases that the algorithm takes a long time.
Once we press OK, the EM algorithm updates the network parameters following the options chosen and comes back with the following dialog:
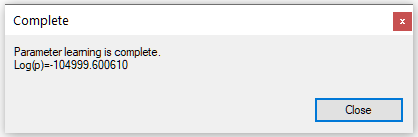
Log(p), decimal logarithm of the probability of data given the structure, also known as log-likelihood, ranging from minus infinity to zero, is a measure of fit of the model to the data.
A remark on the network structure and also on existing evidence. Learning parameters functionality focuses on learning parameters, not the structure, which is assumed fixed and will be unaffected. Existing evidence in the network is ignored and has no effect on the learned parameters.
Finally, a remark on a limitation in learning parameters of continuous, multi-variate Gaussian models. Our implementation of the EM algorithm in this case does not allow for missing values. An extension of this implementation is on our development agenda, so please stay tuned!