Hardware and software requirements
Disk space
Full installation of GeNIe requires less than 50 MB of disk space.
Memory
GeNIe has practically no minimum memory requirements and can run under a minimum Windows configuration. The actual memory requirement will depend on the size and complexity of the models that you create. Too little memory may result in decreased performance. In general, conditional probability tables grow exponentially with the number of parents of a node. The maximum number of parents of a node will, therefore, determine memory requirements. In addition, memory requirements of the clustering algorithm grow with the connectivity of the network.
Operating system
GeNIe has been written for the Windows operating systems. Installation of GeNIe under Windows operating systems may require administrator privileges. While we cannot guarantee 100% compatibility, we are verifying with each build that it runs on macOS (formerly OS X) and Linux under Wine. Please see GeNIe on a Mac and GeNIe on Linux sections for more information on running GeNIe on a macOS and Linux respectively.
Directory, file, and path naming
GeNIe is compatible with any language version of Windows and any Unicode-based naming of files and directories, so directory path names containing non-Latin characters, such as for example Arabic, Chinese, Cyrillic, Hebrew, Hindi, and special diacritical characters in languages otherwise using the Latin alphabet, are not a problem.
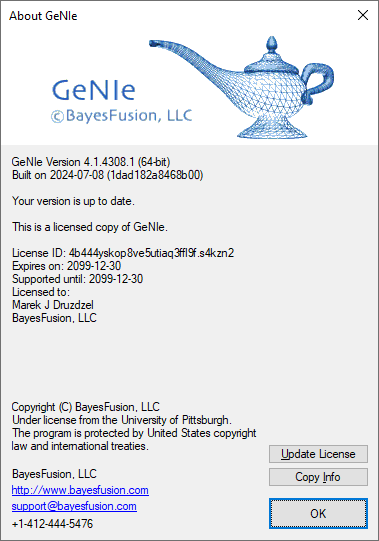
GeNIe version
To determine the version of GeNIe that you have installed, select About GeNIe from the Help Menu. The version number is listed in the small frame of the following window:
Examples
GeNIe installation creates a directory named Examples, containing a sizable library of example models, data sets, and networks used in this manual. The directory has several sub-directories, listed alphabetically below:
Clemen Models: models extracted from the textbook by Robert T. Clemen, Making Hard Decisions: An Introduction to Decision Analysis (Clemen, 1996).
Discrete Bayesian Networks: the most popular type of Bayesian network models, those consisting of discrete variables.
Dynamic Bayesian Networks: discrete-time models used for modeling dynamic systems.
Hybrid Bayesian Networks: models consisting of continuous variables specified by means of equations or mixtures of continuous and discrete variables.
Influence Diagrams: influence diagram models.
Learning: example data sets and other auxiliary files useful in learning Bayesian networks and causal discovery.
Models from the Manual: a collection of models and other auxiliary files used in this manual.
Qualitative Models: qualitative models that can be opened only with QGeNIe.