Clustering algorithm is the fastest known exact algorithm for belief updating in Bayesian networks. It was originally proposed by Lauritzen and Spiegelhalter (1988) and improved by several researchers, e.g., Jensen et al. (1990) or Dawid (1992).
The clustering algorithm works in two phases: (1) compilation of a directed graph into a junction tree, and (2) probability updating in the junction tree. It has been a common practice to compile a network and then perform all operations in the compiled version. Our research in relevance reasoning (Lin & Druzdzel 1997, 1998) has challenged this practice and has shown that it may be advantageous to pre-process the network before transferring it into a junction tree. GeNIe does not include the compilation phase in its user interface and the model under construction is always ready to perform inference.
The clustering algorithm, like all of the algorithms for Bayesian networks, produces marginal probability distributions over all network nodes. In addition, it is possible to preserve clique potentials in the network, which allows for viewing joint probability distribution over those variables that are located within the same clique. Should you wish to derive the joint probability distribution over any variable set, just make sure that they are in the same clique before running the clustering algorithm. One way of making sure that they are in the same clique is creating a dummy node that has all these variables as parents. In any case, when the Preserve Clique Potentials flag is on, there is an additional button in the Value tab of Node Properties dialog, Show JPD (![]() ), which will open a dialog for selecting a set of variables for viewing the joint probability distribution over them.
), which will open a dialog for selecting a set of variables for viewing the joint probability distribution over them.
The clustering algorithm is GeNIe's default algorithm and should be sufficient for most applications. Only when networks become very large and complex, the clustering algorithm may not be fast enough. In that case, we suggest that the user choose an approximate algorithm, such as one of the stochastic sampling algorithms. The best stochastic sampling algorithm available for discrete Bayesian networks is EPIS-BN (Yuan & Druzdzel, 2003).
SMILE's clustering algorithm in UAI-2006 and UAI-2008 inference evaluation
SMILE's implementation of the clustering algorithm underlies the implementation of calculation of the marginal probability and the probability of evidence. SMILE did very well in the UAI-2006 and UAI-2008 inference evaluation. We report the results of the evaluation in the probability of evidence section. Here we show the results of the marginal probability competition in UAI-2008. The following plot show the cumulative time to solve the test instances. The lower the curve, the better. SMILE is shown by the red curve (see https://www.ics.uci.edu/~dechter/softwares/benchmarks/UAI08/uai08-evaluation-2008-09-15.pdf for the full report).
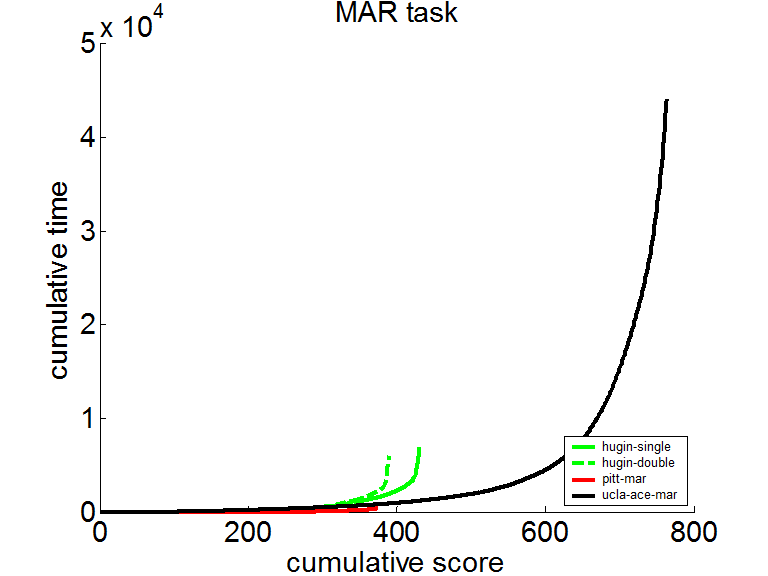
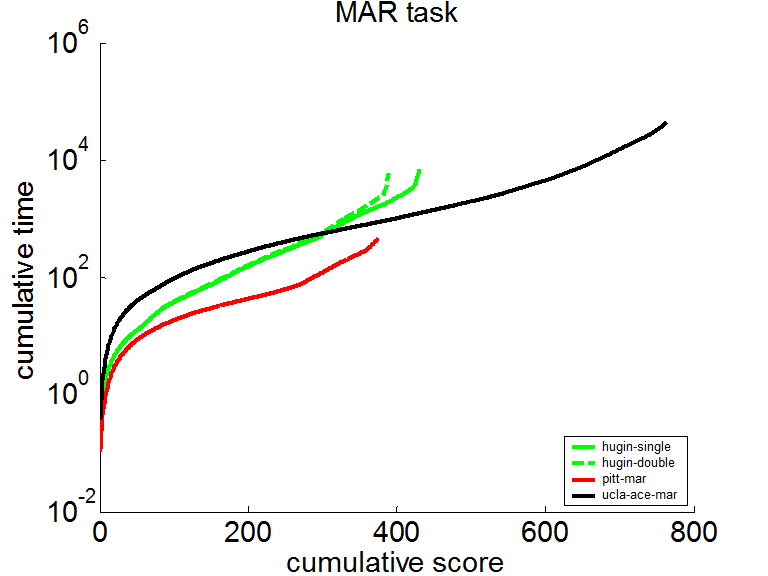
The same curve in logarithmic scale shows that SMILE was typically half an order of magnitude faster than the other software taking part in the competition.
See also the evaluation results for computing the probability of evidence and approximate marginals in the Probability of evidence and EPIS Sampling sections respectively.