Autodiscretization is an approach to solving hybrid Bayesian networks in which there have been observations made in nodes that have parents. In its intermediate stage, the algorithm converts a hybrid Bayesian network into a discrete Bayesian network. For any continuous variable, the user can specify a discretization to follow in this conversion. Once the network has been converted, it behaves like a discrete Bayesian network. The big advantage of this algorithm is that is converts the original hybrid Bayesian network into a discrete Bayesian network only for the purpose of inference, preserving the modeling freedom of the original model specification.
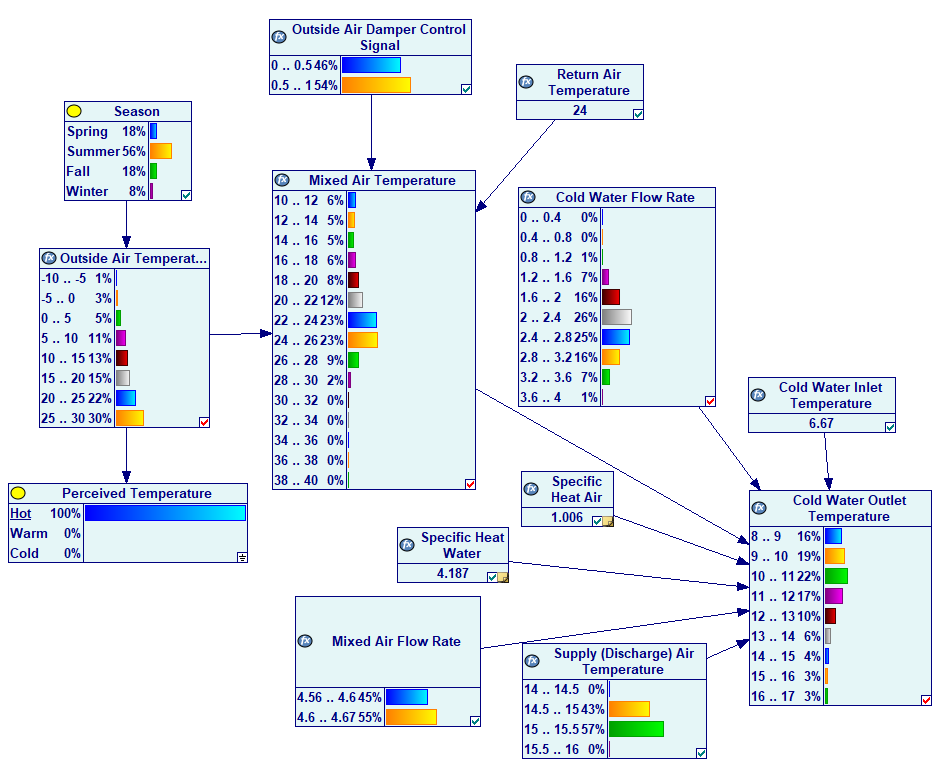
A continuous node in GeNIe is not bound by any constraints and its definition can be a general equation, including any function. Here is the definition of a node Tma (Mixed Air Temperature):
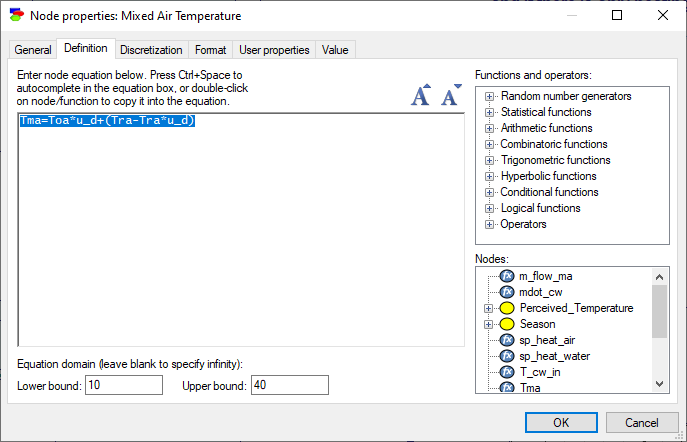
Every continuous node in GeNIe has a Discretization tab. The Discretization tab specifies how the variable should be discretized for the purpose of inference. Here is a possible discretization for the node Tma:
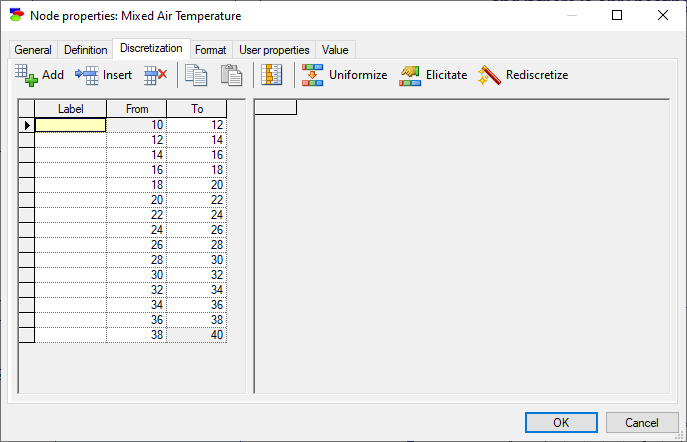
Clicking on the button Rediscretize (![]() ) invokes a simple and local forward sampling algorithm that, based on the discretization of the node and the discretizations of its parents, derives the discrete conditional probability distribution for the node. Here is a distribution derived for the node Tma:
) invokes a simple and local forward sampling algorithm that, based on the discretization of the node and the discretizations of its parents, derives the discrete conditional probability distribution for the node. Here is a distribution derived for the node Tma:
It may happen in the process of auto-discretization that a warning appears in the Output window of the following type:
Discretization problem in node Tma:
Underflow samples: 3000, min=-9.80332 loBound=10
Total valid samples: 13000 of 16000
CPT configs with no valid samples: 1 of 16
Generally, there is no need to worry about this message and the fact that there were underflow or overflow samples has minimal impact on the discretized network. Essentially, when performing discretization, SMILE generates samples from the domains of the parent nodes. Since the child node is specified by an equation, sometimes such samples fall outside of the domain of the child node. This is when underflow/overflow happens. One can observe this in continuous network when the Reject out-of-bound and invalid samples flag is checked (in Network Properties, Inference tab). Sample rejection in forward sampling (both inference in continuous networks and in discretization amounts to forward sampling) is just fine. So, our advice is that when the number of invalid/overflow samples is large, please check if the domain of the variable is not too tight. If the number of invalid/overflow samples is small and the domain bounds are reasonable, we recommend to ignore this message.
Consider the Heat Equations Autodiscretized Hybrid.xdsl network in which an observation has been made in the node Perceived Temperature. Because the node Perceived Temperature has a parent, GeNIe invokes the autodiscretization algorithm. Please note that each of the continuous variables has been discretized. The network behaves like a discrete Bayesian network.