Noisy-MAX is a generalization of the popular canonical gate Noisy-OR that is capable of modeling interactions among variables with multiple states. If all the nodes in question are binary, a Noisy-MAX node reduces to a Noisy-OR node. The Noisy-MAX, as implemented in SMILE, includes an equivalent of negation (through re-ordering of states). By DeMorgan's laws, the OR function (or its generalization, the MAX function) along with a negation, is capable of expressing any logical relationship, including the AND (and its generalization, MIN). This means that SMILE's Noisy-MAX can be used to model the Noisy-AND/MIN functions, as well as other logical relationships.
SMILE's clustering algorithm contains special code path for networks with Noisy-MAX nodes, which can speed up computations significantly. See the Inference section of this manual for details.
To create a Noisy-MAX node, use the DSL_NOISY_MAX type with DSL_network::AddNode:
int h = net.AddNode(DSL_NOISY_MAX, "node1");
AddNode will in this case create a node with a definition object of DSL_noisyMAX class. In addition to information about node outcomes, the definition contains the following Noisy-MAX specific data:
•a set of conditional probabilities, also called outcome strengths
•for each of parent node, the vector of parent outcome strengths.
The conditional probabilities are stored in a two-dimensional DSL_Dmatrix object, the Noisy-MAX table, accessible through DSL_noisyMAX::GetCiWeights. The size of the first dimension of the matrix is the sum of the number of each parent outcomes plus one (for the leak column). The second dimension is the number of the child node's own outcomes. The last column for each of the parents is constrained to contain zero probabilities in all but its last element.
Each set of parent outcomes can be ordered specifically for the current interaction with the child node. The Noisy-MAX table always follows the specified order of parent outcomes.
As an example, consider a binary Noisy-MAX node with two parents, each with three outcomes. The following snippet modifies the probabilities and outcome strengths for the second parent (with zero-based index of 1).
auto maxDef = net.GetNode(h)->Def<DSL_noisyMAX>());
DSL_Dmatrix weights = maxDef->GetCiWeights();
int BASE = 2 * 3;
weights[BASE] = 0.1;
weights[BASE + 1] = 0.9;
weights[BASE + 2] = 0.3;
weights[BASE + 3] = 0.7;
maxDef->SetCiWeights(weights);
DSL_intArray strengths({2, 0, 1});
maxDef->SetParentOutcomeStrengths(1, strengths);
The value of BASE is calculated as a product of node outcome count and preceding parents' outcome counts (in this case, there is just one preceding parent with three outcomes). The probabilities for the parent are written into a DSL_Dmatrix object initialized with GetCiWeights, and written back with SetCiWeights. The next step changes the order of parents' outcomes in relationship to the Noisy-MAX (child) node. The DSL_intArray object is created and initialized with parent outcome indices. After the DSL_noisyMAX::SetParentOutcomeStrengths call, the first column of probabilities for the parent with index 1 (the one with 0.1 and 0.9) represents the probabilities for the parent outcome with index 2 (because 2 is the first element in the strengths array). Note that this does not modify the parent node in any way and the ordering is valid only in the context of this particular parent-child relationship. Assuming that we started with the default uniform probabilities in the table, our modifications yields the following Noisy-MAX definition, as viewed in GeNIe:
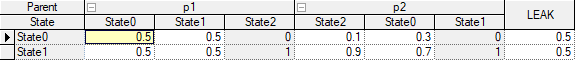
The outcomes of both parents are {State0, State1 and State2}. However, by using DSL_noisyMAX::SetParentOutcomeStrenghts for parent p2, its outcomes are seen by the Noisy-MAX child node as {State2, State0, State1}. Other Noisy-MAX nodes in the same network can set up their own parent outcome ordering if p2 becomes their parent.
See the DSL_noisyMAX reference for details.